raxmlGUI
2.0.5Perform phylogenetic analysis more easy and convenient by taking advantage of the new features in RAxML that are echoed in this simple GUI
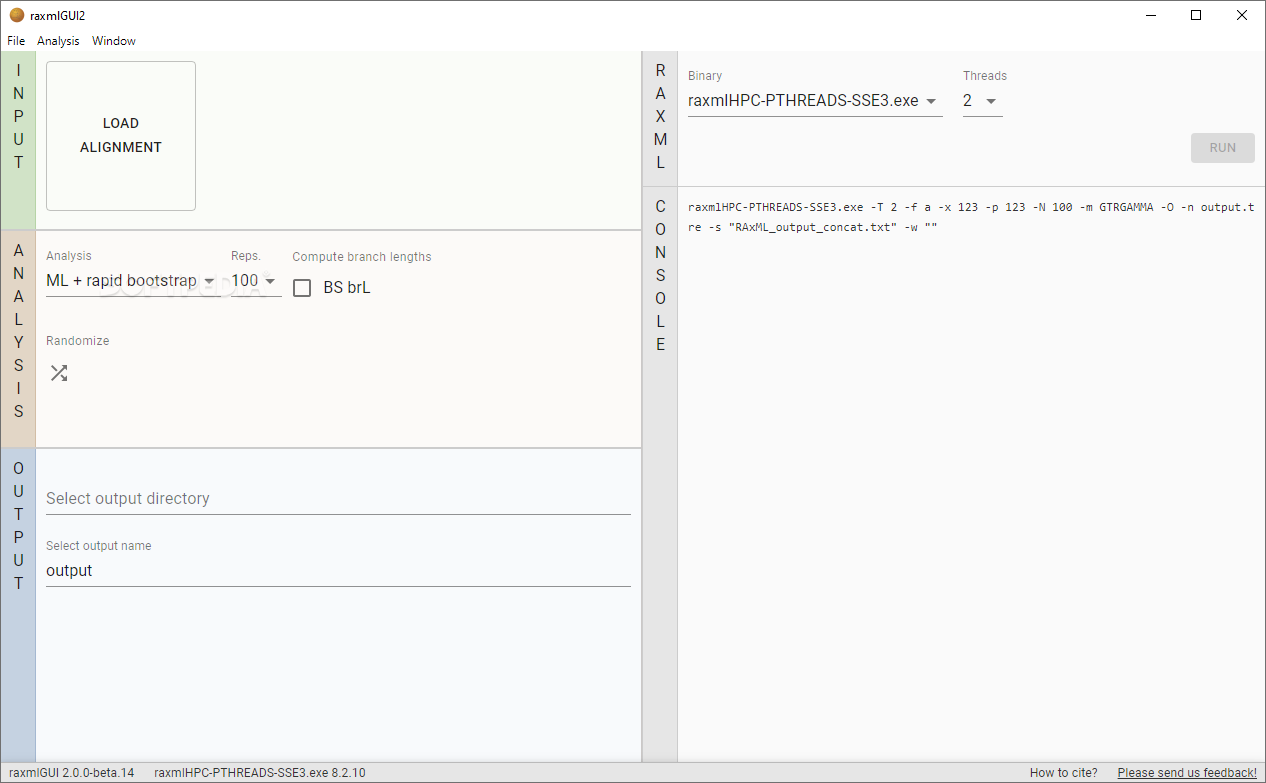
raxmlGUI is a user-friendly, easy-to-use graphical front-end specially designed to help you with phylogenetic analysis using RAxML or Randomized Axelerated Maximum Likelihood. The latter program is widely used for performing phylogenetic analyses, including post-analysis of the sets, alignments or the evolutionary placement of short reads.
The program is updated to support the new features of RAxML, such as bootstrapping and support values as well as the models and data types with binary and multi-state morphological and RNA secondary structure data. The other new features supported include a fine-grain parallelization of the likelihood function, post-analysis of trees, analyzing next generation sequencing data and vector intrinsics that can accelerate parsimony and likelihood calculations.
According to the developer, the Beta version is capable of generating automated pipelines for analyses that require multiple successive calls of RAxML. Therefore, the phylogenetic analyses are simplified thanks to the intuitive interface and the recent updates to RAxML.
The input formats supported by the application include both FASTA and PHYLIP formats. As anyone in the field would argue, the PHYLIP format reserves the first ten characters before each sequence for the name. While it is true that this type of format allows longer names, it also requires no space within the name to know where it ends.
The program is updated to support the new features of RAxML, such as bootstrapping and support values as well as the models and data types with binary and multi-state morphological and RNA secondary structure data. The other new features supported include a fine-grain parallelization of the likelihood function, post-analysis of trees, analyzing next generation sequencing data and vector intrinsics that can accelerate parsimony and likelihood calculations.
According to the developer, the Beta version is capable of generating automated pipelines for analyses that require multiple successive calls of RAxML. Therefore, the phylogenetic analyses are simplified thanks to the intuitive interface and the recent updates to RAxML.
The input formats supported by the application include both FASTA and PHYLIP formats. As anyone in the field would argue, the PHYLIP format reserves the first ten characters before each sequence for the name. While it is true that this type of format allows longer names, it also requires no space within the name to know where it ends.
92.8 MB
Info
Update Date
May 08 2021
Version
2.0.5
License
AGPL
Created By
Daniele Silvestro & Ingo Michalak
Related software CAD